The single most significant biological event since creation is undoubtedly the Curse on creation as a result of Adam's sin. In addition to animal death, creationists have long inferred a number of other effects of the Curse, including biological poisons,1 parasites,2 predation,3 and microbial pathogens.4 Creationists generally explain these Curse-related imperfections as degenerations of originally beneficial structures.5, 6 The modern genomics revolution in biology is producing confirmatory evidence of this creationist idea. (Briefly defined, a genome can be thought of as the complete genetic complement of an organism, including every nucleotide of DNA from every chromosome. Of the 39 genomes that have been completely sequenced at the time of this writing, 21 [54%] are microbial pathogens.)
In a recent review, Wren7 discusses three possible origins for bacterial pathogenicity, our understanding of which has been greatly expanded by genomic research:
- Lateral gene transfer in which bacteria transfer genes for pathogenicity or antibiotic resistance from one cell to another.
- Antigenic variation in which the bacteria have active mechanisms that produce varying types of antigens in order to elude the host defense systems.
- Genome decay in which the genomes of pathogenic organisms lose many important genes that are no longer essential in the host environment.
Of these three themes, genome decay is most consistent with the creationist idea of a degenerating creation. A discussion of lateral gene transfer and antigenic variation is beyond the scope of this article; however, lateral gene transfer in pathogenicity has been reviewed elsewhere.8, 9
The best documented example of pathogen-associated genome decay is found in the group of bacteria known as mycoplasmas. The mycoplasmas are unique in that they lack an outer membrane and a cell wall,10 and they also use an atypical genetic code.11 Taken together, these two characteristics strongly suggest a phylogenetic discontinuity with other bacteria. The mycoplasmas may therefore be labeled an apobaramin.12 Because bacteria reproduce asexually, determining the boundaries of the holobaramins of the mycoplasmas is far more difficult than in sexually reproducing eukaryotes where the hybridization criterion may be employed; however, evidence discussed below indicates the likelihood that at least two species of mycoplasmas, Mycoplasma genitalium and M. pneumoniae are members of the same monobaramin.
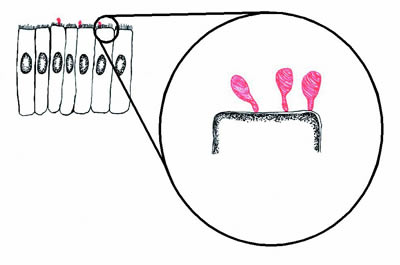 |
|
Figure 1. Diagram of mycoplasmas (red) infecting epithelial cells (black). The mycoplasmas form a small structure called a "tip" by which they adhere to the epithelial cells. If the mycoplasmas are prevented from adhering to the epithelial cells, they are avirulent. |
The complete genomes of three mycoplasmas have been published, M. genitalium, M. pneumoniae, and Ureaplasma urealyticum, all of which are mucosal pathogens of humans. M. genitalium is found in the urinary tract and is associated with non-gonococcal urethritis. Its genome contains 580,070 nucleotides and 468 genes and is the smallest known genome of any self-reproducing organism.13 M. pneumoniae causes atypical pneumonia and has a slightly larger genome of 816,394 nucleotides and 677 genes.14 U. urealyticum also inhabits the human urinary tract and is an opportunistic pathogen during human pregnancy. Its genome contains 751,719 nucleotides and 613 genes.15 All 468 genes of M. genitalium are also found in the genome of M. pneumoniae,16 indicating a high probability that these two bacterial species are members of the same monobaramin. The relationship of U. urealyticum to these two species is much more questionable. Only 324 U. urealyticum genes (53% of its gene complement) are found in either M. genitalium or M. pneumoniae;17 thus, the baraminic status of U. urealyticum cannot be assigned with any confidence.
An important feature of the genomes of M. genitalium and M. pneumoniae is the genes that they lack. Although both species retain the ability to synthesize proteins via translation, neither has the capability of synthesizing amino acids, the building blocks of proteins.18 Thus, all amino acids must be obtained from the host via transport across the mycoplasma cell inner membrane. The absence of important biosynthetic genes is believed to be a hallmark of genome decay. But how do we know whether the created ancestors of M. genitalium and M. pneumoniae had the ability to synthesize amino acids? Could the lack of amino acid synthesis genes be a design feature of this baramin? To answer this, we must identify a mechanism of gene loss.
If, as it appears, M. genitalium and M. pneumoniae are members of the same monobaramin, they share a common ancestor. Any genomic differences between the two species are the result of changes that have occurred after their divergence. As mentioned above, every M. genitalium gene is also found in M. pneumoniae, but the genome of M. pneumoniae is larger than the M. genitalium genome by an additional 209 genes. The 468 genes shared between M. genitalium and M. pneumoniae are arranged in six contiguous segments bounded by highly similar repetitive sequences. Within the six segments, the gene order is nearly identical, but the segments themselves are arranged differently in each genome. Because the segments are bounded by repetitive sequences, it is very probable that the differences in the two genomes were caused by faulty recombination. Recombination in bacterial genomes is accomplished by the RecA protein, and the gene for RecA is found in both M. genitalium and M. pneumoniae.19 Considering the propensity for genomic rearrangement, it is possible that small or even large segments of genes could be lost by faulty recombination. Because these two species of mycoplasmas share a common ancestor and because a mechanism for gene loss is present, it is almost certain that true genome decay has occurred in this monobaramin.
In the evolutionary model, pathogenicity and parasitism is thought to progress from very virulent (aggressive) forms to harmless or even mutually beneficial relationships. Advocates claim that natural selection will favor hosts that are resistant to the parasite and parasites that are not rapid killers of their own host environments. Thus, as time progresses, the parasites evolve to less virulent forms, and the hosts become tolerant of the more benign forms of the parasites.20 In the case of the mycoplasmas, I propose the opposite interpretation. Rather than starting out as a virulent relationship, the human/mycoplasma relationship may have been harmless or beneficial at creation. In this interpretation, important genes were lost from the mycoplasma as time progressed, rendering the mycoplasmas increasingly dependent on their hosts for survival. Thus, the pathogenicity of these organisms may be an indirect consequence of the loss of important genetic information. This interpretation is remarkably consistent with the traditional creationist understanding of a now degenerating world.
Glossary
Baramin: created kind.
Holobaramin: A group containing all and only organisms related by common descent.
Monobaramin: A group containing only organisms related by common descent, but not necessarily all of them.
Apobaramin: Any group of holobaramins that are separated from all other organisms by phylogenetic discontinuities.
References
- Batten, D., ed., The Revised & Expanded Answers Book (Green Forest, AR: Master Books, 1990), p. 103.
- Armitage, M.H., "Trematode Parasites: What is their Genesis?" Creation Research Society Quarterly 36 (2000): 184-194.
- Stambaugh, J., "Creation's Original Diet and the Changes at the Fall," Creation Ex Nihilo Technical Journal 5 (1991): 130-138.
- Morris, H.M., The Genesis Record (San Diego, CA: Creation-Life Publishers, 1976), pp. 125-126.
- Morris, ref. 4, p. 125.
- McCoy, D.L., "Evidence Against the Evolutionary Superiority of Plant Pathogenesis," in Proceedings of the 1992 Twin-Cities Creation Conference (Roseville, MN: Twin-Cities Creation Science Association, 1992), pp. 186-191.
- Wren, B.W., "Microbial Genome Analysis: Insights into Virulence, Host Adaptation, and Evolution," Nature Reviews Genetics 1 (2000): 30-39.
- Penrose, E., "Bacterial Resistance to Antibiotics…A Case of Un-Natural Selection," Creation Research Society Quarterly 35 (1998): 76-83.
- Wood, T.C., "The AGEing Process: Rapid Post-Flood Intrabaraminic Diversification Caused by Altruistic Genetic Elements (AGEs)," Origins (2001), submitted.
- Woese, C.R., J. Maniloff, and L.B. Zablen, "Phylogenetic Analysis of the Mycoplasmas," Proceedings of the National Academy of Sciences USA 77 (1980): 494-498.
- Osawa, S., T.H. Jukes, K. Watanabe, and A. Muto, "Recent Evidence for Evolution of the Genetic Code," Microbiology Reviews 56 (1992): 229-264.
- ReMine, W.J., "Discontinuity Systematics: A New Methodology of Biosystematics Relevant to the Creation Model," in Proceedings of the Second International Conference on Creationism, vol. 2, edited by R.E. Walsh and C.L. Brooks (Pittsburgh, PA: Creation Science Fellowship, 1990), pp. 207-213.
- Fraser, C.M. and 28 others, "The Minimal Gene Complement of Mycoplasma pneumoniae," Science 270 (1995): 397-403.
- Himmelreich, R., H. Hilbert, H. Plages, E. Pirkl, B.-C. Li, and R. Herrmann, "Complete Sequence Analysis of the Genome of the Bacterium Mycoplasma pneumoniae," Nucleic Acids Research 24 (1996): 4420-4449.
- Glass, J.I., E.J. Lefkowitz, J.S. Glass, C.R. Heiner, E.Y. Chen, and G.H. Cassell, "The complete sequence of the mucosal pathogen Ureaplasma urealyticum," Nature 407 (2000): 757-762.
- Himmelreich, R., H. Plagens, H. Hilbert, B. Reiner, and R. Herrmann, "Comparative analysis of the genomes of the Bacteria Mycoplasma pneumoniae and Mycoplasma genitalium," Nucleic Acids Research 25 (1997): 701-712.
- Glass, et al., ref. 15.
- Himmelreich, et al., ref. 17.
- Himmelreich, et al., ref. 17.
- Pianka, E.R., Evolutionary Ecology (New York, NY: Harper Collins, 1988), p. 296.
* Dr. Wood is Assistant Professor at the Center for Origins Research, Bryan College, Tennessee and received his Ph.D. in Genetics from Clemson University.
Cite this article: Wood, T. C. 2001. Genome Decay in the Mycoplasmas. Acts & Facts. 30 (10).











































